Key Takeaways
- The Human Genome Project (HGP) mapped all the genes in the human genome, significantly advancing our understanding of human genetics.
- Comparative genomics, part of the HGP, compared human genes with those of other species, such as the mouse and platypus, to gain insights into our genetic similarities, evolutionary history and unique human traits.
- The HGP has profoundly impacted medical research, particularly in understanding and treating cancer, by enabling the sequencing of cancer genomes and fostering personalized medicine approaches based on genetic mutations.
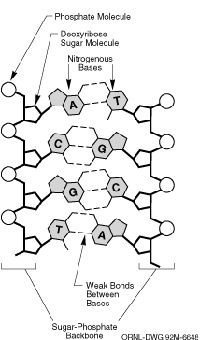
Space may be the final frontier, but human biology is the original unknown, challenging us to discover who we are and where we came from. DNA, the building block of life, contains the genetic code that informs so much of who we are. This code is written with four letters, each representing a different base. The four bases are adenine (A), which pairs with thymine (T), and cytosine (C), which pairs with guanine (G).
Scientists have long known that these four letters provide the recipes for proteins, which carry out numerous bodily functions. But there are still questions to be answered, including how the 3.2 billion base pairs contained in the human genome are ordered. (The human genome is a person's entire bundle of DNA divided unevenly among 23 pairs of chromosomes.) To that end, the Human Genome Project (HGP) was launched in 1990. Some of the project's ambitious goals included:
Advertisement
- Sequencing the entire human genome
- Identifying human genes
- Charting variations across human genomes
- Sequencing genomes of the mouse and four other "model organisms"
[source: Genome.gov]
Run by the National Institutes of Health and the U.S. Department of Energy, the project was completed ahead of schedule in 2003. A "final" batch of results was published in 2006, but data produced by the HGP are continually examined, analyzed and occasionally revised. Theoretically, with the main goals achieved, the project is finished. Let's look at some of what we learned.
Only a few years before the completion of the HGP, popular predictions stated that humans had up to 100,000 genes. But recent HGP estimates lowered that number to a more modest range of 20,000 to 25,000 [source: Human Genome Project Information]. In addition, the HGP has helped to narrow the range of possible genes and to isolate certain candidates as contributing to specific diseases. Scientists have also reassessed previous assumptions, such as the idea that genes are self-contained, discrete pieces of DNA with defined roles. That's not always the case. We now know that some multitasking genes make more than one protein; in fact, the average gene may make three proteins [source: Genome.gov]. Also, genes appear to grab genetic code from other DNA segments.
Before we look closely at heredity and genes, let's stop to consider what scientists have learned about animal and other genomes. Some of these projects, such as mapping the mouse genome, were included in the original Human Genome Project and can tell us about our evolution and DNA.
Advertisement